NEW YORK – Your brain is made up of nerve cells that come in an incredible array of shapes, sizes, and functions, working together to grant you the ability to think, move and sense the world. Yet every one of these cells contains exactly the same genes: about 20,000. How is this possible? How can the same set of genes be used to generate so many different kinds of nerve cells in the brain?
Using fruit fly embryos, the lab of Minoree Kohwi, PhD, Zuckerman Institute principal investigator, has found one piece of the puzzle. Her latest research, which examines how neurons form, highlights the importance of the physical location of genes within the nucleus of the neuroblast, the stem cell from which the neurons are born. We sat down to talk with Dr. Kohwi, also an assistant professor of neuroscience at Columbia’s Vagelos College of Physicians & Surgeons, and Tanguy Lucas, PhD, a postdoctoral research fellow in her lab, about their new discoveries on the link between genome architecture and neural diversity, published September 27 in Developmental Cell.
What is genome architecture, and why is it important for the development of an organism?
Minoree Kohwi: High school textbooks tend to depict the DNA contained inside a cell’s nucleus as being jumbled, with genes, which are stretches of DNA that encode the protein building blocks of our cells, seemingly distributed at random. But the last decade or so has seen an explosion in the field of genome architecture, the study of how our genome is organized and packaged within the nucleus, fueled by recent advances in DNA sequencing and mapping. We are now getting a better picture of how the nucleus arranges and distributes genes in an orderly fashion, kind of like books in a library. This organization is thought to play a crucial role in orchestrating which set of genes are turned on or off to create different types of cells.
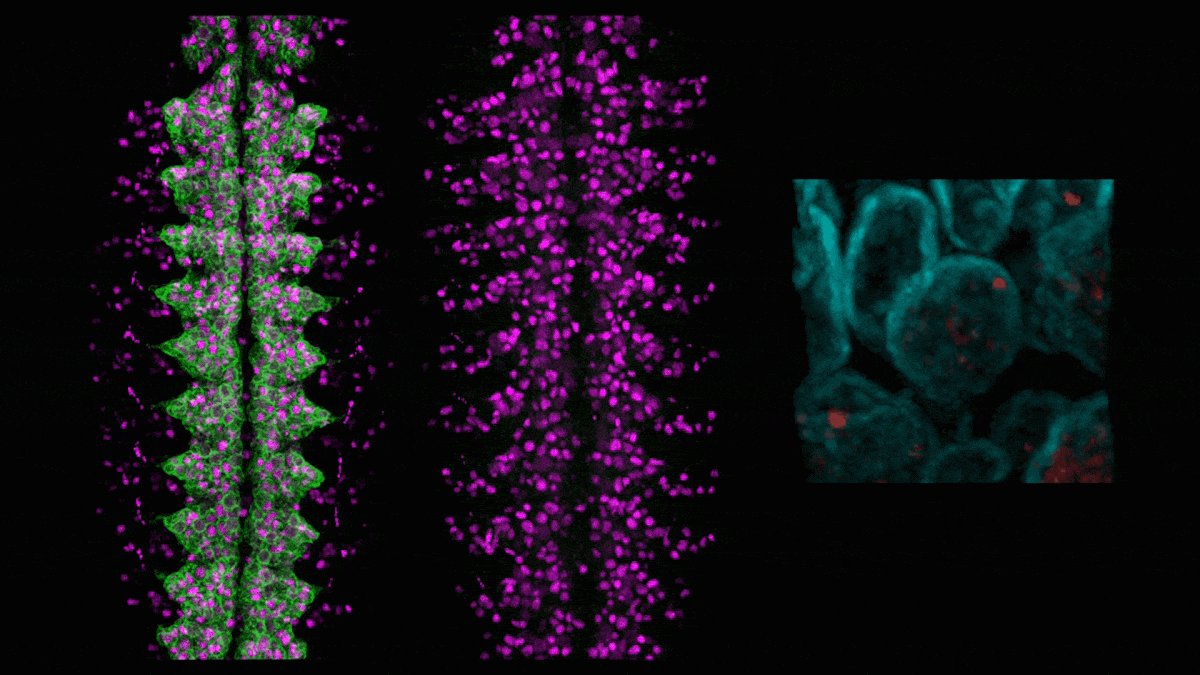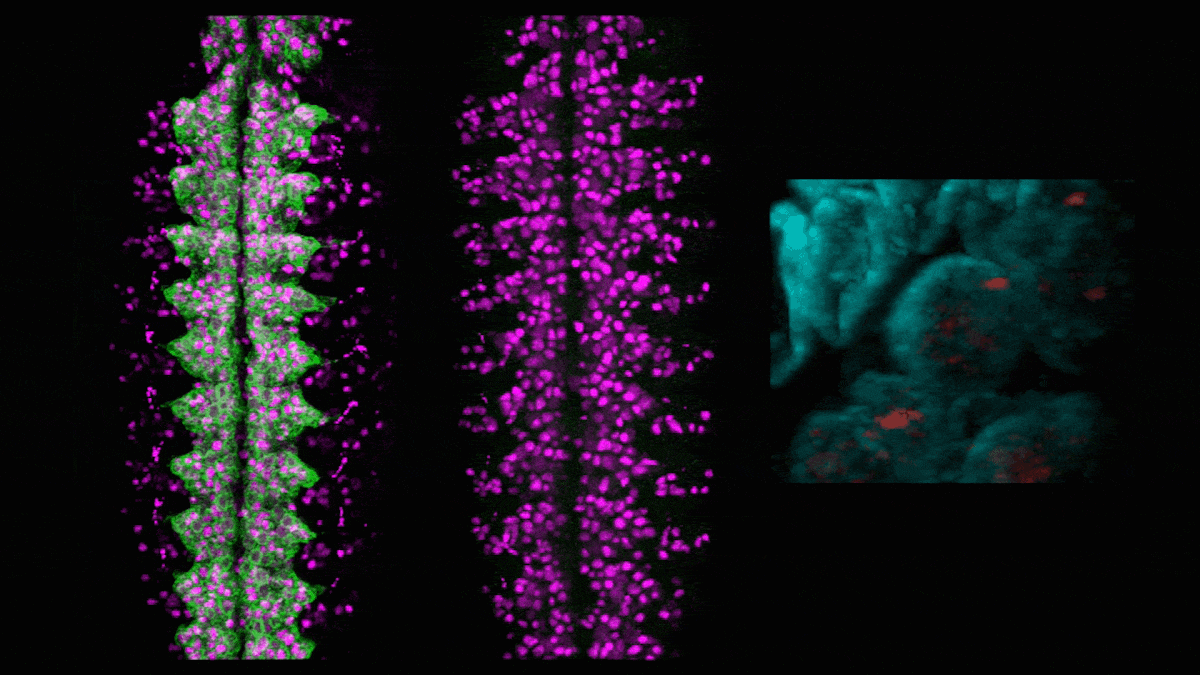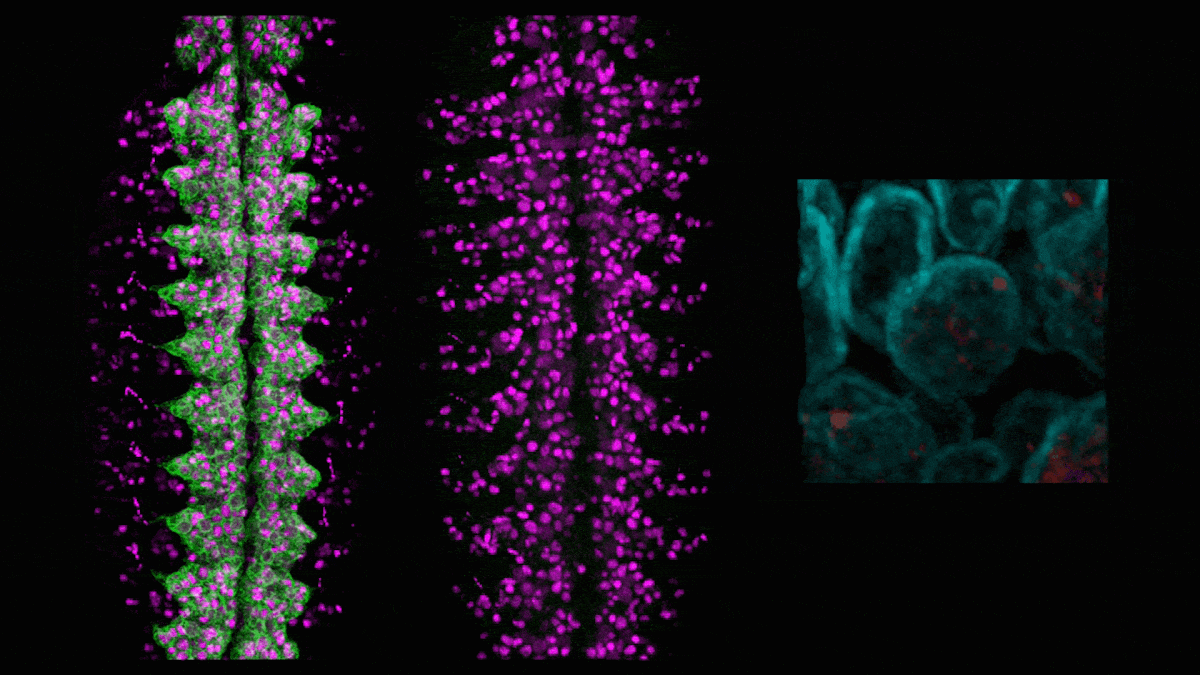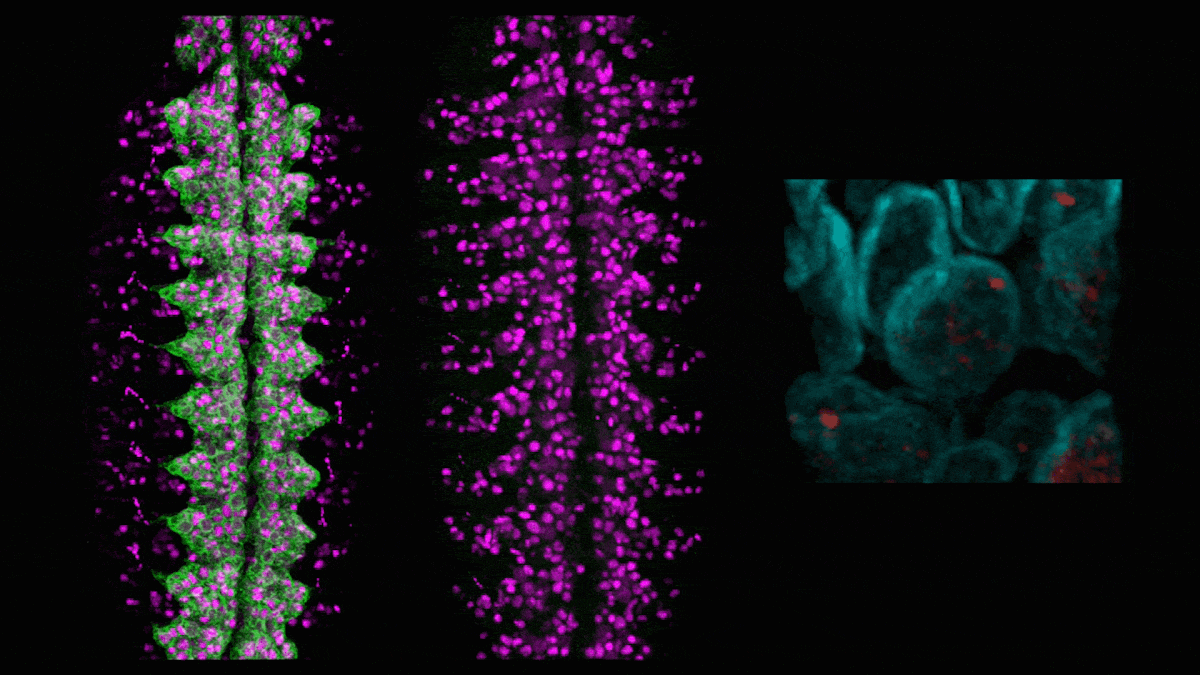
In neuroblasts (purple) in a developing fly embryo, a gene for green fluorescent protein (left) is silenced (middle) by adding the hunchback intron, which causes the gene (red, right) to relocate to the nuclear lamin (cyan). (Credit: Tanguy Lucas/Kohwi Lab/Columbia's Zuckerman Institute)
So the physical location of genes in the nucleus has an effect on whether they get turned on or off, during the course of an embryo’s development.
Kohwi: Exactly. We also know that genes can move within the nucleus, which changes how they behave. My lab has been exploring how this movement is controlled in neural stem cells. Is there a code embedded within genes themselves? What proteins interact with genes to facilitate relocation? Studying the fruit fly allows us to visualize and track individual genes and address these questions in intact, developing animal. We also have a lot of tools for manipulating a fly’s genetics to reveal the mechanisms involved in gene relocation. These are things you can’t easily do in other research model systems, and at the basic level, these biological concepts apply to us humans as well.
Tanguy Lucas: We started our investigation with the nuclear lamina, a fibrous mesh at the edge of the nucleus, where the cell stores away genes it doesn’t want expressed. For a cell to maintain its identity, it’s important not just to control what genes it turns on, but what genes it keeps off. For a neuron that can live as long as the human it inhabits, making sure it’s expressing only the correct set of genes to do its job properly is really important. Scientists know that genes at the lamina tend to be tightly packed, making it difficult for them to be turned on by proteins. To understand how genes wind up at the lamina, we focused on a gene called hunchback.
As in the Hunchback of Notre Dame?
Kohwi: Fly biologists have a thing for playful names. The name hunchback comes from the fly’s hunched appearance when the gene is mutated.
What do we know about hunchback?
Kohwi: As a fly nervous system develops, the dividing stem cells, called neuroblasts, make a lot of different neurons; hunchback turns on in the very first of those neurons made. After a limited number of divisions neuroblasts lose the ability to make these early neurons. Our previous work found that this is due to the hunchback gene moving to the nuclear lamina in the neuroblasts, where it becomes permanently stored away.
Lucas: In our current experiments, we took at closer look at hunchback in neuroblasts isolated from fly embryos. We found that inside the gene was a piece of DNA not as tightly packed as we would expect it to be at the lamina. It was the intron, which no one had paid much attention to as they don’t encode for the protein parts. Introns are fascinating, quiet DNA elements involved in almost all steps between reading the gene’s instructions to the production of the proteins they encode. But the function of introns in regulating genome architecture remains unknown. In our system, we showed that getting rid of part of this intron stopped hunchback from relocating. Attaching the intron to other bits of DNA caused them to relocate to the nuclear lamina.
So the intron helps move the DNA?
Lucas: Our experiments suggest that the intron is needed to shuttle hunchback to the lamina. Once there, the gene stays put – even after the cell divides. In every new cell, the gene remains stuck and turned off, and that turned-off state is inherited by the daughter neurons that are born from the neuroblasts. What’s particularly exciting is that we found that Polycomb proteins seem to attach to the intron region and are necessary for the hunchback gene to relocate to the nuclear lamina. Polycomb proteins have long been studied for their role in preventing genes from turning on in the wrong place and time (in flies and humans alike), but controlling gene movement to the nuclear lamina was a new function for these proteins. So, in summary, we found a hidden DNA code that allows the cell to permanently store the hunchback gene, after it has done its job and been switched off, and we found the proteins that facilitate this movement.
The last decade or so has seen an explosion in the field of genome architecture.
So how does this process of genome reorganization fit into the bigger picture of brain development?
Kohwi: Similar things might be happening to lots of other genes – some are sent away for permanent storage, while others might be coming out of storage when they’re needed at subsequent stages of brain development. Genes, and the proteins they encode, are like ingredients for a particular type of cell. In my kitchen, I can use the same pots and pans to cook lots of different meals, depending on what’s available in my fridge. Reorganizing the genome is like organizing the limited fridge space so that only the relevant ingredients are available for cooking and using the same pots and pans (the molecular machinery) to create different dishes (cells). And storing away certain ingredients might be a way to prevent the wrong ingredient from ending up in your dish, salt instead of sugar, for example, which could be catastrophic. Evolutionarily, this would be an efficient way to increase the variety of cells without having to exponentially increase the size of the genome. Case in point, fruit flies and humans have roughly the same number of genes, but we humans have a lot more cell diversity.
Lucas: Studying how the genome is organized and how it interacts with the physical structures of the nucleus like the lamina, could tell us how neurodevelopmental diseases arise. It could also give us insights into issues associated with aging. Studies have shown, for instance, that disruptions in the nuclear envelope can contribute to neural degeneration associated with diseases like Alzheimer’s. We’re part of the explosion of research now looking into all of these questions, and we’re excited to find out more.
###
Read more about their new work in “Discrete cis-acting element regulates developmentally timed gene-lamina relocation and neural progenitor competence in vivo,” published September 27 in Developmental Cell. Authors on the paper are Tanguy Lucas, Terry L. Hafer, Harrison Zhang, Natalia Molotkova and Minoree Kohwi.