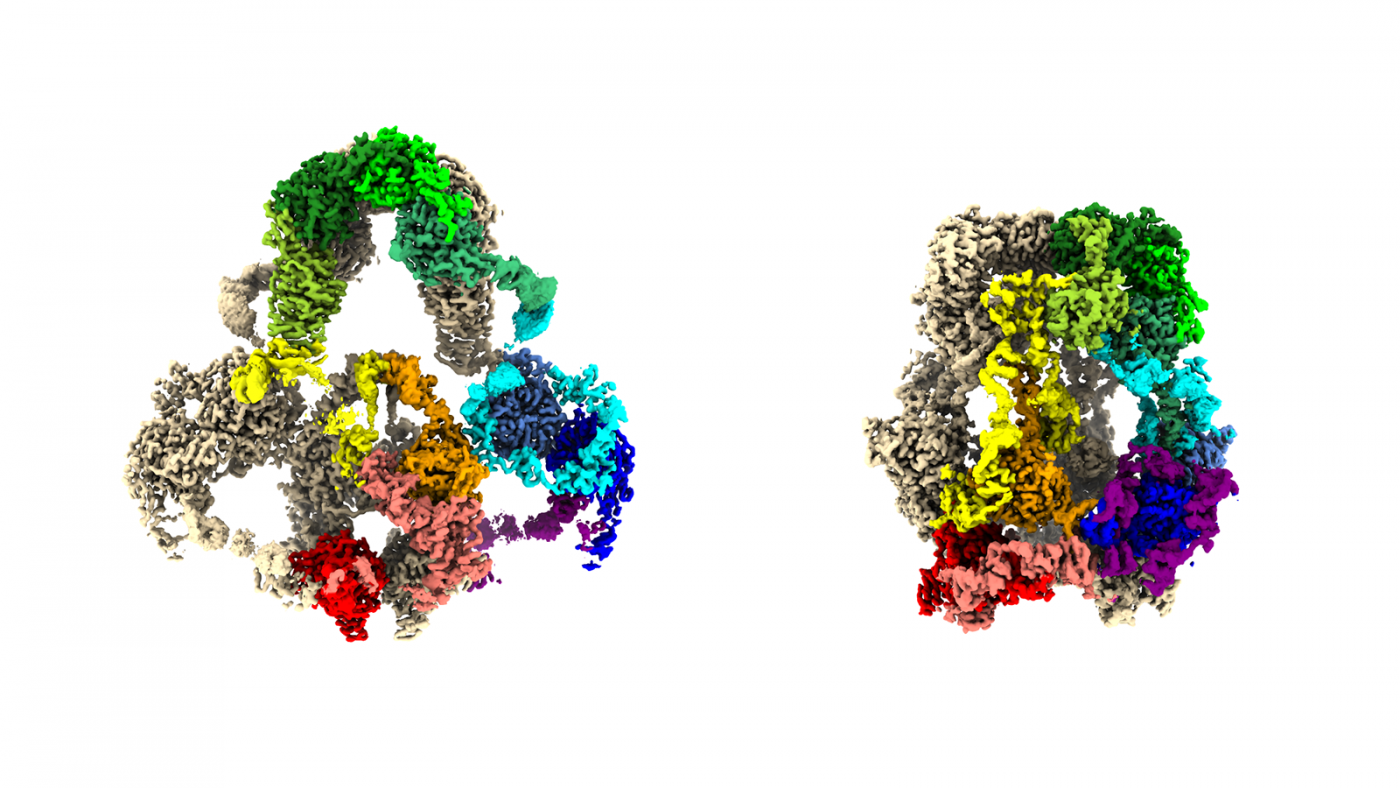NEW YORK, NY — The devil so often is in the details. For proteins that orchestrate the molecular business of life, there are devils and angels in their details, down to the proteins’ constituent atoms. It’s at that level of structural minutiae where the balance of health and disease, even life and death, can pivot.
Published today online in the journal Cell, a collaboration of nephrologists and neuroscientists at Columbia University showed the value that emerges from uncommon alliances. They and colleagues elsewhere reveal for the first time a portrait of a life-and-death protein with enough clarity to finally reveal how it works: as a minuscule ferry for molecular passengers that must cross nearly a trillion cell membranes in tissues and organs ranging from kidneys and brains to the inner ear and the lungs’ alveoli.
“With new mechanistic understanding of this key protein, and how mutations in it can shut it down, we are hoping that follow-on research will uncover novel targets for treating kidney and brain diseases,” said Jonathan Barasch, MD, PhD, an expert and clinician in urology and nephrology at Columbia’s Vagelos College of Physicians and Surgeons and a corresponding author on the paper. "These new therapeutic openings are due to the amazing protein structures my Columbia colleagues Andrew Beenken, Anthony Fitzpatrick and Larry Shapiro have uncovered."
Dr. Barasch and his coauthors envision that the new high-resolution protein structures will point toward therapeutic leads for treating diseases as prevalent as acute kidney injury (affecting more than 4 million patients per year in the USA), chronic kidney disease (affecting some 800 million people worldwide) and Alzheimer’s disease (affecting an estimated 32 million people globally), and as rare as Donnai-Barrow syndrome (affecting fewer than 1,000 Americans), a genetic disorder with multiple physical and cognitive consequences.
The protein is known as LRP2, a member of a family of LRP proteins found in creatures ranging from worms to people. Compared to most proteins on cell membranes, LRPs are huge, so much so that the scientists who discovered LRP2 in the early 1980s, Marilyn Farquhar and Dontscho Kerjaschki, dubbed it megalin. Some LRPs are built from more than 4,600 amino acids, the molecular building blocks of all proteins.
In kidney cells, LRP2 is crucial for recovering reusable molecules from filtered metabolic wastes from bodily fluids so the body does not have to spend energy and resources to make them again. For each of these cells, there are likely tens of thousands of LRP2 proteins, distributed on the surface like seeds on a strawberry.
“The kidney is faced with recovering 99 percent of the body’s salt and water that passes through the organ’s filters, and also with recovering 100 percent of small proteins that otherwise would dump into the urine and out of the body,” said Dr. Barasch. “There have been general ideas about how this recovery works, but its specificity has now been solved.”
We now have the best 3D maps of the LRP2 protein ever created
This is where Dr. Barasch’s colleagues come in. One of them is Andrew Beenken, MD, PhD, a biochemist and nephrologist at Columbia and the paper’s first author. Membrane proteins like those in the LRP family are notoriously difficult to isolate, let alone map out in detail. Dr. Beenken navigated a giant step around that impasse with an arduous process in which he deployed a bevy of biochemical techniques.
“When I first heard about what Jonathan and Andrew were planning to do, I did not think it would be possible,” said co-corresponding author Lawrence Shapiro, PhD, a principal investigator at the Zuckerman Institute and a professor of biochemistry and molecular biophysics at Columbia’s Vagelos College of Physicians and Surgeons. Dr. Shapiro’s expertise in teasing out how a protein’s complex structure begets its biological functions would prove crucial in deciphering LRP2’s ferrying mechanism.
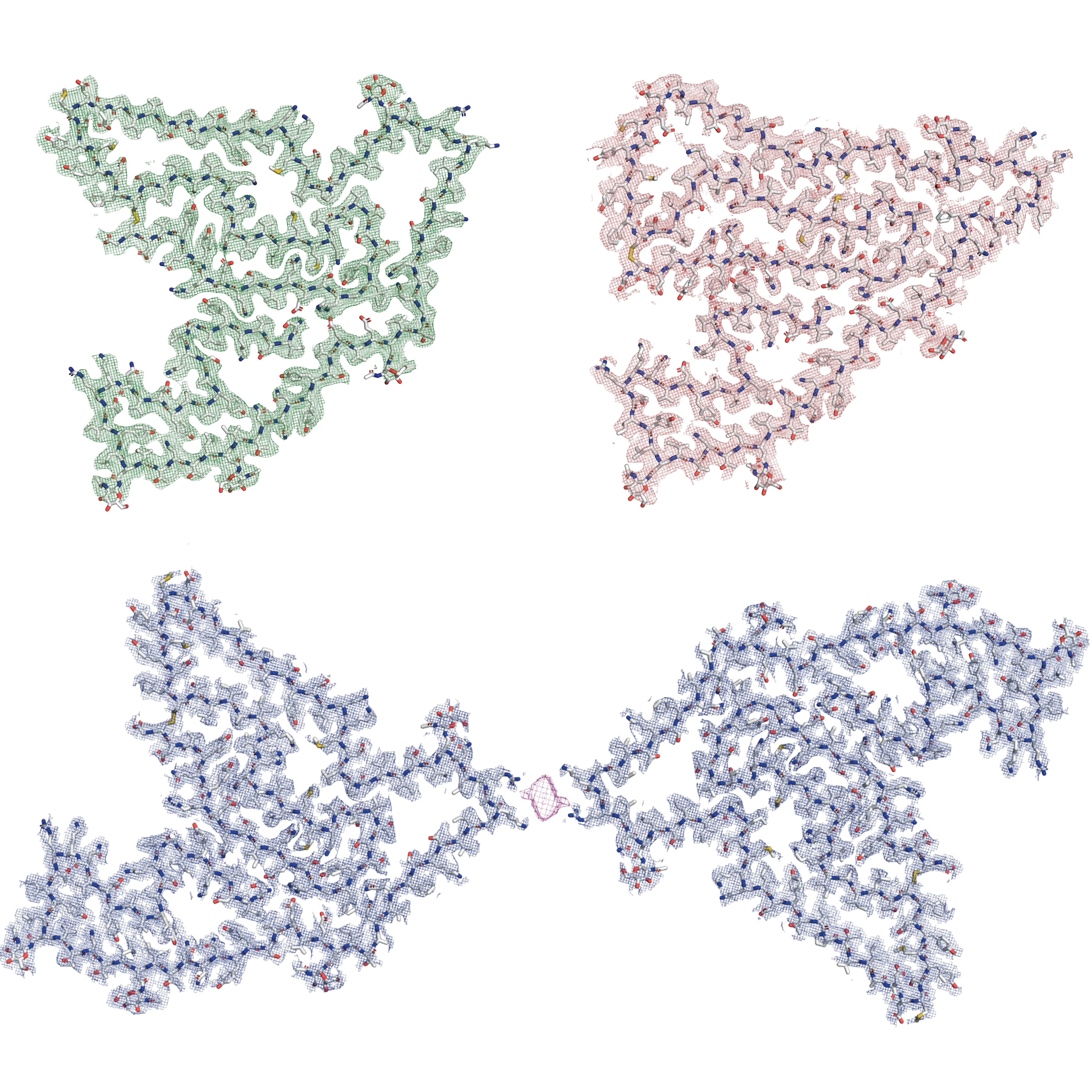
With a mix of benchtop skill, creativity and determination, Dr. Beenken harvested enough LRP2 protein from 500 mouse kidneys to solidify the protein into a sample of sufficient size and purity for analysis with advanced microscopy techniques. In his harvesting of the LRP2 molecules, Dr. Beenken pulled off a biochemical tour de force: capturing LRP2 proteins locked into two of their key conformations, a crucial laboratory feat for unveiling the protein’s machine-like actions in cells.
This is where a second giant research step came in, this one led by co-corresponding author Anthony Fitzpatrick, PhD, a leader in the field of cryogenic electron microscopy, which is especially suited for studying large proteins and other biomolecules.
With a two-story, liquid-nitrogen-cooled, cryogenic electron microscope, Dr. Fitzpatrick and Dr. Beenken collected vast amounts of structural data using Dr. Beenken’s hard-won LRP2 samples. Then, with the deft use of powerful computational tools to make sense of the data, the researchers produced 3D protein structures in near-atomic detail.
“We now have the best 3D maps of the LRP2 protein ever created,” said Dr. Fitzpatrick, a principal investigator at the Zuckerman Institute and an assistant professor of biochemistry and molecular biophysics at Columbia’s Vagelos College of Physicians and Surgeons. With those maps, Dr. Shapiro could begin to tease out the remarkable mechanism by which LRP2 works in cells.
An analysis of cryo-electron microscope images revealed how the enormous protein megalin (LRP2) changes shape (A. Fitzpatrick, A Beenken, L. Shapiro / Columbia's Zuckerman Institute).
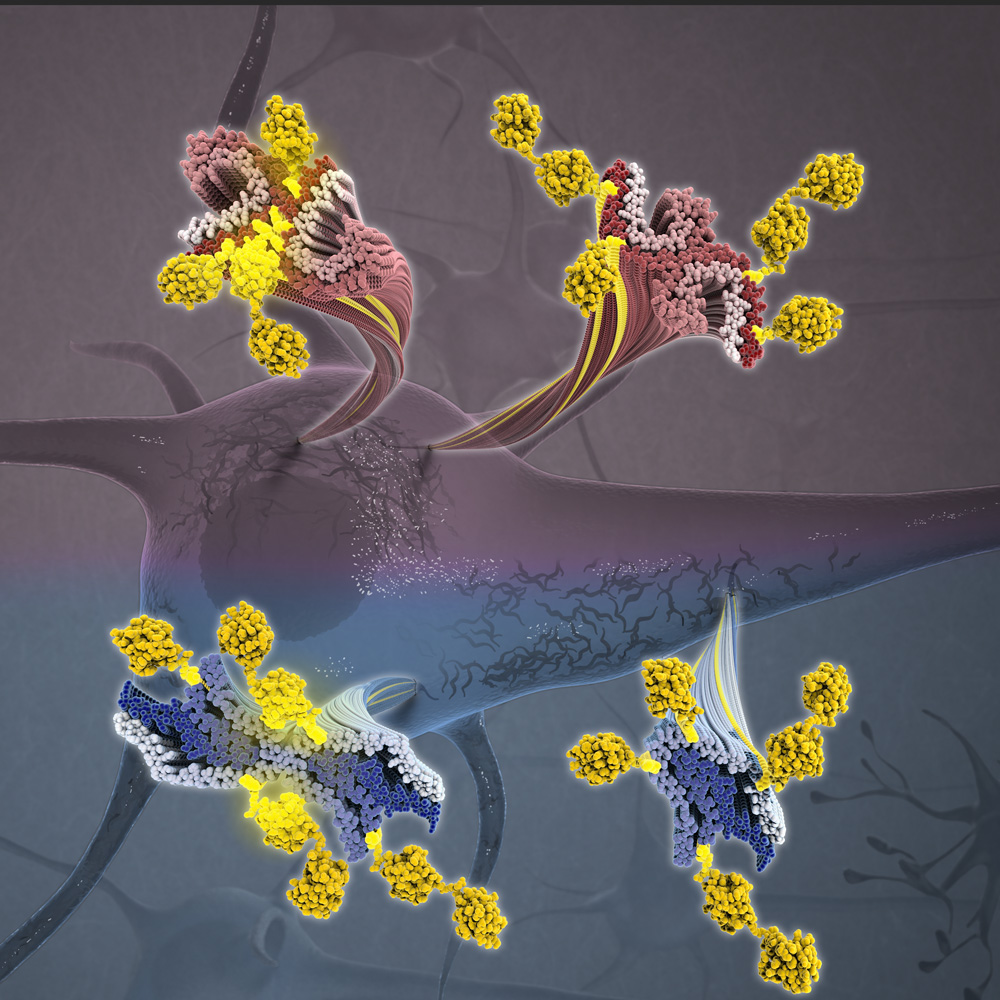
One of the mapped-out conformations captures the shape of the LRP2 when it resides on and within a cell membrane. That’s where the protein picks up molecular passengers from the liquid outside the cell–whether from the urine produced in the kidney or the liquid around brain cells. Among those passengers are small proteins, including tau and amyloid-beta (both implicated in Alzheimer’s disease), insulin and ones that shuttle vitamins A and D around cells.
The other LRP2 conformation is the one the protein snaps into after it becomes enveloped in a bit of cell membrane and ferries off to locations inside the cell. It is in these little capsules, endosomes, where the shape-shifting protein either enables its molecular passengers to be recycled intact for further use or to be deconstructed into reusable or disposable components.
From this pair of LRP2 structures, the team was able to determine that the protein undergoes a machine-like toggle between its passenger-embarking form and its passenger-disembarking form. When LRP2 proteins are mutation-free, they succeed in maintaining molecular balances in, for example, blood and brain tissue. But when there are even tiny tweaks in LRPs’ enormous molecular anatomy, these proteins can contribute to disease.
In kidney cells, for one, faulty LRP2 proteins renege on their normal task of retrieving proteins that would otherwise be lost in the urine. That can lead to a variety of conditions including chronic kidney disease, Donnai-Barrow syndrome and conditions that are lethal in neonatal stages.
In the brain, LRP2 (and the related LRP1) normally help clear a variety of toxins, among them tau protein fragments, which have long been associated with Alzheimer’s disease. But proteins of the LRP family also have been shown to transfer such fragments between brain cells, potentially contributing to the disease process.
“You could imagine that trying to inhibit this from happening with a drug could be helpful,” said Dr. Shapiro.
This is where the power of cryogenic-EM comes in strong.
“If you know exactly the atomic details of where the tau binds, you might actually block that using either an antibody or a small molecule,” said Dr. Fitzpatrick. “Cryo-EM can get you to the level of detail you need in order to work on potential new therapies.”
“This is the beginning of a long road of discovery of how these LRP proteins work and to new drug targets for a range of diseases,” said Dr. Beenken.
###
The paper, titled “Structures of LRP2 reveal a molecular machine for endocytosis,” was published online in Cell on February 6, 2023. Its full list of authors includes: Andrew Beenken, Gabriele Cerutti, Julia Brasch, Yicheng Guo, Zizhang Sheng, Hediye Erdjument-Bromage, Zainab Aziz, Shelief Y Robbins-Juarez, Estefania Y Chavez, Goran Ahlsen, Phinikoula S. Katsamba, Thomas A. Neubert, Anthony W.P. Fitzpatrick, Jonathan Barasch, Lawrence Shapiro.
Sources of funding and support include grants from the Simons Foundation, NYSTAR, NIH National Institute of General Medical Sciences, Agouron Institute, NIH, NIH NIDDK, NIH NCRR, and NIH NINDS.
The authors report no conflicts of interest.