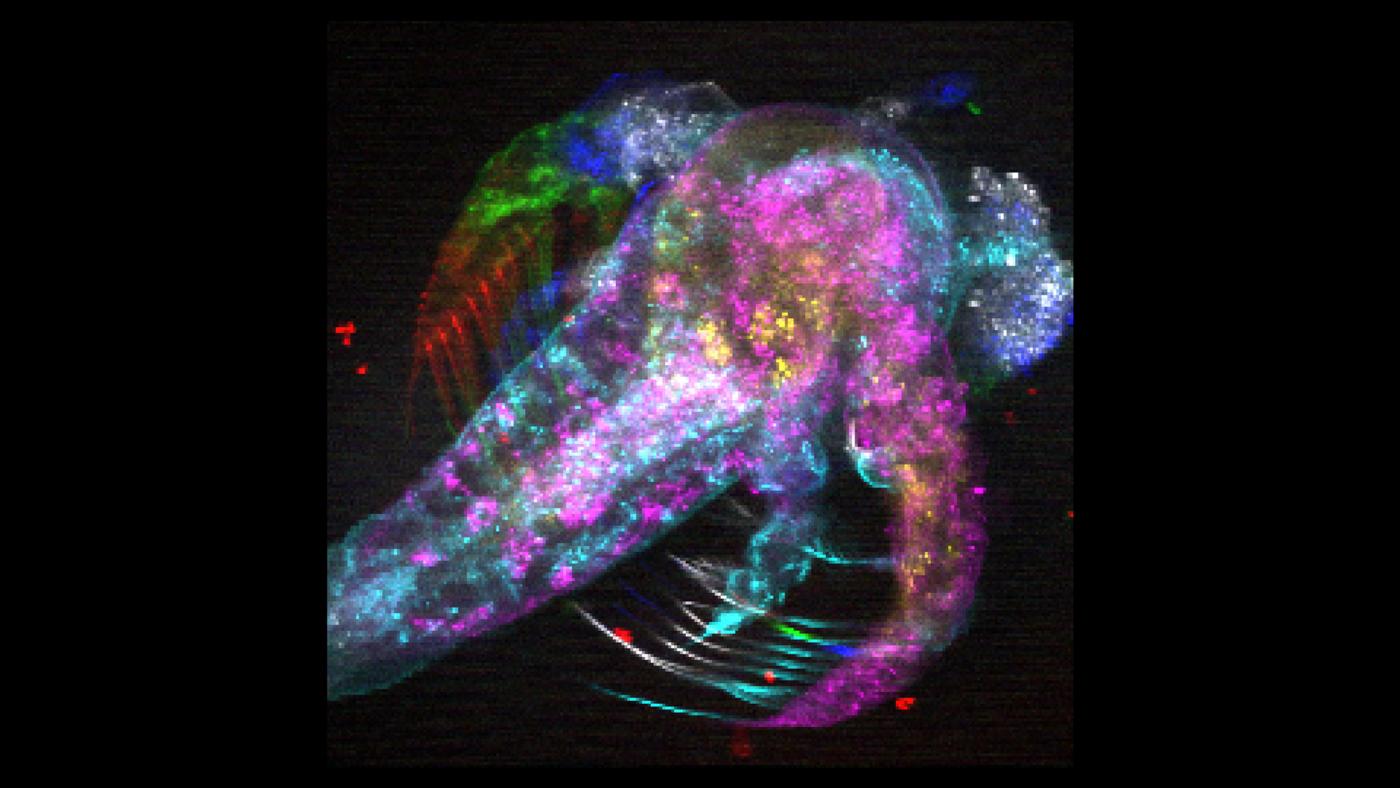
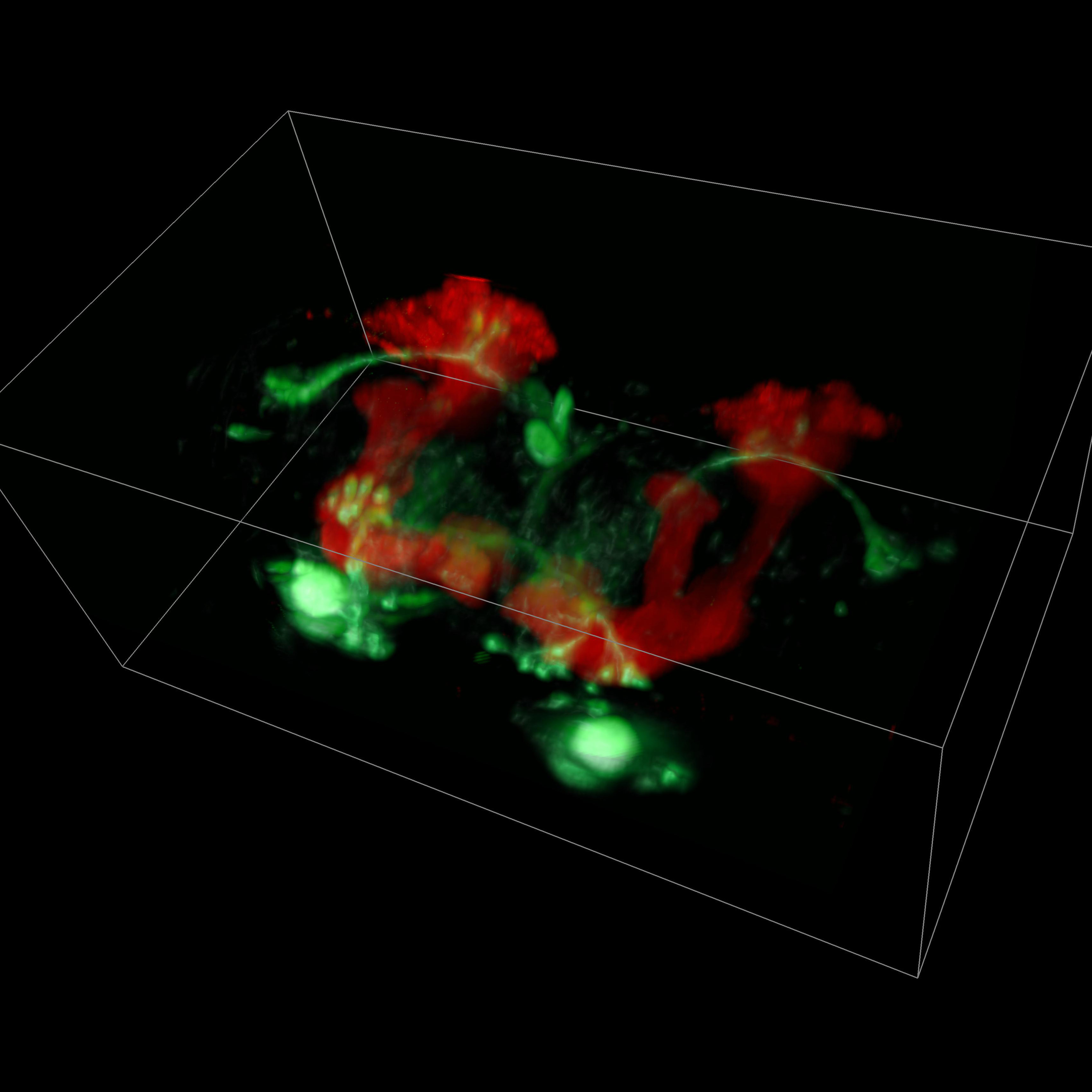
Despite extraordinary advances in technology, obtaining high-resolution, detailed, meaningful images of the brain that shed light on how we move, think, act and feel remains challenging.
Several years ago, a team of neuroscientists in Rafael Yuste’s Lab at Columbia University had become particularly dissatisfied with the widely used, yet functionally problematic, two-photon microscope. So they built a better one, the details of which were published recently in a series of papers in the journal Neuron.
We spoke with three of the papers’ authors: Rafael Yuste; MD, PhD, Columbia professor of biology and neuroscience; Darcy Peterka, PhD, a research associate in the Yuste Lab who recently joined the Mortimer B. Zuckerman Mind Brain Behavior Institute as Director of Cellular Imaging; and Liam Paninski, PhD, professor of statistics at Columbia and a principal investigator at the Zuckerman Institute.
The three scientists discussed how they hacked the two-photon microscope and made sense of the images they captured. They also share what this technological advance could mean for the field of neuroscience.
Building a Better Microscope
How do neuroscientists traditionally image brain cells?
Rafael Yuste: One of the fundamental problems with imaging brain cells is that, for most organisms, the brain is opaque. So rather than looking into a clear piece of glass, you’re peering through a dense fog. Any light that you try to shine through ends up scattering. When I was at Bell Labs in 1995, Winfried Denk (now at the Max Planck Institute of Neurobiology) and I published a paper in Nature that showed you could cut through this fog using two-photon microscopy.
Darcy Peterka: It represented a significant advance in the field, but did come with some limitations. For example, in order to image a small area of the brain, the laser has to be moved to the next spot, over and over again one pixel at a time. Scientists would then piece together a composite image. This is how a standard two-photon microscope works. While this microscope is used in hundreds of labs around the country, it isn’t fast enough to give us a complete picture. To truly understand the brain, you have to be able see — in real time — how cells communicate with each other to produce a thought, a memory or a movement. But this wasn’t possible with the two-photon microscope. It would be like trying to watch a movie — but with every other frame missing. So we wanted to improve upon it.
What were some of the improvements?
DP: One of the initial improvements was to the microscope hardware itself. In a 2008 paper, Volodymir Nikolenko, a graduate student in the Yuste lab, added a spatial light modulator, or SLM, which basically allows you to flexibly split a single light beam into multiple ‘beamlets.’ We could then shine these beamlets at different places around the brain — and use a camera to capture all the images simultaneously. So instead of imaging one pixel at a time, we could image many.
RY: This improvement enabled a faster, more panoramic view of the brain — especially for taking wide shots — but it couldn’t image very deeply. For deep brain imaging, scattering turned everything into a complicated mess. You could tell something was happening in the deeper layers, but you couldn’t clearly see what it was.
When did you start looking into the problem?
RY: Since the 2008 paper, our lab began focusing on coupling spatial light modulators with two-photon microscopes. At the time, we weren’t able to use this modification to perform the kind of deep imaging we had planned on. But we reasoned there should be a way to do it; we just had to figure out how.
DP: For me, I’ve always enjoyed pulling things apart, tweaking them, and then putting them back together in new ways — a skill that has proven particularly useful for the field of experimental neuroscience. The original SLM microscope was powerful, but it just couldn’t do everything we needed.
What alterations did you make?
DP: First we ripped apart the old system and rebuilt it — but with improvements. By putting the spatial light modulator and the light source together in just the right way, we could then very quickly and accurately project to any position in three-dimensional space, simply by running some software. We then combined this improved ability to target with the microscope’s ability to scan, creating a hybrid microscope that combined the best features of both methods. And with the SLM in place, we had the backdoor we needed to image deeper into the brain.
RY: The SLM multi-beam hack essentially gave us a more complete composite image of the brain as a starting point. But in order to make sense of the image we had captured, we turned to statistician and data scientist Liam Paninski.
Finding Signals in the Noise
Dr. Paninski, you are a trained statistician. What can statistics bring to the field of neuroscience?
Liam Paninski: The sheer amount of data that neuroscientists can now generate is staggering. A single experiment can yield a full terabyte of data — which is the equivalent of the average computer’s entire hard drive. As imaging techniques continue to improve, that number will continue to skyrocket.
As a result, they partner with people like me — statisticians, data scientists and theorists — who can help to make sense of the vast amounts of data they generate. As we move forward, I foresee the partnerships between neuroscientists and data scientists only building in strength as we attempt to unravel the mysteries of the brain and mind.
How did this most recent collaboration with the neuroscience labs originate?
LP: It’s been clear for a long time that two-photon imaging is a very powerful method, used in hundreds of labs around the world, so we’ve been interested in figuring out the best way of extracting the data in an ordered and meaningful way. When the Yuste Lab came to us with imaging data from their modified microscope, we saw this as a great opportunity to develop that method.
One way of looking at the work from the Yuste lab is that they have these very nice tricks for modifying the microscope to mash multiple images together into one composite image. But if you don’t have a good way of then separating out those images for analysis, then everything that has been done is for naught.
Our work highlights the importance of what the neuroscientists have done — mashing the images together to speed things up — but also our ability to separate it all out in a systematic way in order to make sense of what was captured.
The whole game here, given the current limitations on cameras, is that we must come up with some way of quickly observing many cells simultaneously, and then rely on data science to make sense of it.
How do you hope to use these approaches — both the SLM and the statistical approach — in future studies?
LP: The goal is to come as close to real-time analysis as possible. In an ideal world, we’d be able to pull the images off the microscope and do the analysis right then and there. If you can identify what you’re seeing in real time, then you can manipulate or control it.
RY: In fact, in some cases we’re getting close. In the future, we foresee the systematic mapping of brain activity — at the cellular level — in three dimensions, as a key to truly understanding the brain and mind. This is exactly the central idea behind the Brain Activity Map, a proposal for a large-scale technology development project to build better tools to read and write neuronal activity in neural circuits. This proposal has inspired the White House’s BRAIN Initiative. Tools such as this one will be developed that will hopefully let us finally crack the neural code.
###
These papers are titled:
“Simultaneous multi-plane imaging of neural circuits.” Authors include: Weijian Yang, Jae-eun, Kang Miller, Luis Carrillo-Reid, Eftychios Pnevmatikakis, Liam Paninski, Rafael Yuste and Darcy S. Peterka.
"Simultaneous denoising, deconvolution, and demixing of calcium imaging data." Authors include: Eftychios A. Pnevmatikakis, Daniel Soudry, Yuanjun Gao, Timothy A. Machado, Josh Merel, David Pfau, Thomas Reardon, Yu Mu, Clay Lacefield, Weijian Yang, Misha Ahrens, Randy Bruno, Thomas M. Jessell, Darcy S. Peterka, Rafael Yuste and Liam Paninski.